HEALPIX images¶
Images can also be stored using the HEALPIX representation, and the
reproject package includes two functions,
reproject_from_healpix() and
reproject_to_healpix(), which can be used to reproject
from/to HEALPIX representations (these functions are wrappers around
functionality provided by the astropy-healpix
package). These functions do the reprojection using interpolation (and the
order can be specified using the order argument). The functions can be
imported with:
from reproject import reproject_from_healpix, reproject_to_healpix
The reproject_from_healpix() function takes either a
filename, a FITS Table HDU object, or a tuple containing a 1-D array and a
coordinate frame given as an Astropy BaseCoordinateFrame
instance or a string. The target
projection should be given either as a WCS object (which required you to also
specify the output shape using shape_out) or as a FITS
Header object.
To demonstrate these functions, we can download an example HEALPIX map which is a posterior probability distribution map from the LIGO project:
from astropy.utils.data import get_pkg_data_filename
filename_ligo = get_pkg_data_filename('allsky/ligo_simulated.fits.gz')
We can then read in this dataset using Astropy (note that we access HDU 1 because HEALPIX data is stored as a binary table which cannot be in HDU 0):
from astropy.io import fits
hdu_ligo = fits.open(filename_ligo)[1]
We now define a header using the Mollweide projection:
target_header = fits.Header.fromstring("""
NAXIS = 2
NAXIS1 = 480
NAXIS2 = 240
CTYPE1 = 'RA---MOL'
CRPIX1 = 240.5
CRVAL1 = 180.0
CDELT1 = -0.675
CUNIT1 = 'deg '
CTYPE2 = 'DEC--MOL'
CRPIX2 = 120.5
CRVAL2 = 0.0
CDELT2 = 0.675
CUNIT2 = 'deg '
COORDSYS= 'icrs '
""", sep='\n')
All of the following are examples of valid ways of reprojecting the HEALPIX LIGO data onto the Mollweide projection:
With an input filename and a target header:
array, footprint = reproject_from_healpix(filename_ligo, target_header)
With an input filename and a target wcs and shape:
from astropy.wcs import WCS target_wcs = WCS(target_header) array, footprint = reproject_from_healpix(filename_ligo, target_wcs, shape_out=(240,480))
With an input array (and associated coordinate system as a string) and a target header:
data = hdu_ligo.data['PROB'] array, footprint = reproject_from_healpix((data, 'icrs'), target_header, nested=True)
Note that in this case we have to be careful to specify whether the pixels
are in nested (nested=True) or ring (nested=False) order.
With an input array (and associated coordinate system) and a target header:
from astropy.coordinates import FK5 array, footprint = reproject_from_healpix((data, FK5(equinox='J2010')), target_header, nested=True)
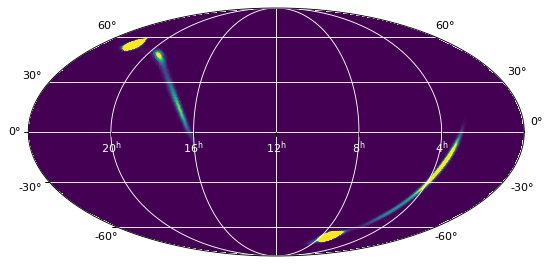
The resulting map is the following:
from astropy.wcs import WCS
import matplotlib.pyplot as plt
from astropy.visualization.wcsaxes.frame import EllipticalFrame
ax = plt.subplot(1,1,1, projection=WCS(target_header),
frame_class=EllipticalFrame)
ax.imshow(array, vmin=0, vmax=1.e-8)
ax.coords.grid(color='white')
ax.coords['ra'].set_ticklabel(color='white')
On the other hand, the reproject_to_healpix() function takes
input data in the same form as reproject_interp()
(see Interpolation) for the first argument, and a coordinate frame as the
second argument, either as a string or as a
BaseCoordinateFrame instance e.g.:
array, footprint = reproject_to_healpix((array, target_header), 'galactic', nside=128)
The array returned is a 1-D array which can be stored in a HEALPIX FITS file.
We can use the Table object to easily write the array to a
HEALPix FITS file:
from astropy.table import Table
t = Table()
t['flux'] = array
t.meta['ORDERING'] = 'RING'
t.meta['COORDSYS'] = 'G'
t.meta['NSIDE'] = 128
t.meta['INDXSCHM'] = 'IMPLICIT'
t.write('healpix_map.fits')
Note
When converting to a HEALPIX array, it is important to be aware
that the order of the array matters (nested or ring). The
reproject_to_healpix() function takes a nested
argument that defaults to False, hence why we set ORDERING to
'RING'.